如何使用MLflow️简化ML的工作流程
简介
MLflow 是一个开源平台,可帮助 ML 工程师和数据科学家管理从实验和开发到部署和监控的整个机器学习生命周期。它提供了用于跟踪实验、打包代码、共享模型并将其部署到生产环境中的工具。它简化了构建和使用机器学习模型的过程,使跟踪进度、复制结果以及将模型部署到实际应用中变得更加容易。它还为管理这些任务提供了一个集中式界面以及可视化工具。
我们将构建一个葡萄酒质量预测预测模型,深入实际地研究它。
葡萄酒质量预测E2E工作流程
项目结构
在这里,我使用了 cookie-cutter 模板来创建标准化的项目结构。
日期收集
我将数据集保存在 Google Drive 上,因此下面我实现了从驱动器中提取数据并将其保存到本地存储的代码。数据集可在此处获取:
import pathlib
import yaml
import pandas as pd
from src.logger import infologger
infologger.info('*** Executing: load_dataset.py ***')
# load data from given path and return df
def load_data(remote_loc: str) -> pd.DataFrame :
try :
# correct way to read data from drive
remote_loc = 'https://drive.google.com/uc?id=' + remote_loc.split('/')[-2]
df = pd.read_csv(remote_loc)
except Exception as e :
infologger.info(f'there\'s some issue while loading data from remote server [check load_data()]. exc: {e}')
else :
infologger.info(f'data loaded from {remote_loc}')
return df
# save data at data/raw dir
def save_data(data: pd.DateOffset, output_path: str, file_name: str) -> None :
try :
data.to_csv(path_or_buf = output_path + f'/{file_name}.csv', index = False)
except Exception as e :
infologger.info(f'there\'s some issue while saving the data [check save_data()]. exc: {e}')
else :
infologger.info(f'data saved at [path: {output_path}/{file_name}.csv]')
# load data & then save it
def main() -> None :
curr_dir = pathlib.Path(__file__)
home_dir = curr_dir.parent.parent.parent
params_file = home_dir.as_posix() + '/params.yaml'
try :
params = yaml.safe_load(open(params_file))
except Exception as e :
infologger.info(f'there\'s some issue while loading the params file or output path [check main()]. exc: {e}')
else :
# create dir if not present, else execute without any warning/error
output_path = home_dir.as_posix() + params['load_dataset']['raw_data']
pathlib.Path(output_path).mkdir(parents = True, exist_ok = True)
data = load_data(params['load_dataset']['drive_link'])
save_data(data, output_path = output_path, file_name = params['load_dataset']['file_name'])
infologger.info('program terminated normally!')
if __name__ == "__main__" :
infologger.info('load_dataset.py as __main__')
main()
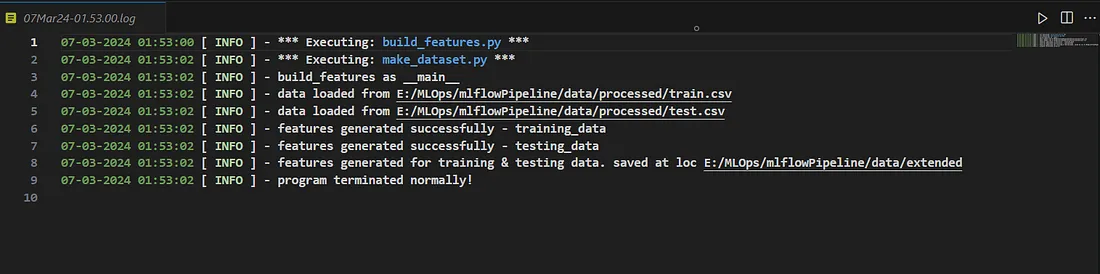
实施记录仪
你可能会注意到,我使用了 infologger 来记录详细信息。我编写了一段代码,为每次执行创建日志文件并存储日志。下面是代码和日志文件的快照,以便更好地理解。
# file: logger.py
import logging
import pathlib
from datetime import datetime
infologger = logging.getLogger(__name__)
infologger.setLevel(logging.INFO)
formatter = logging.Formatter('%(asctime)s : %(levelname)s : %(name)s : %(message)s')
log_file = f'{datetime.now().strftime("%d%b%y-%H.%M.%S")}.log'
# get current path and goto root
log_dir_path = (pathlib.Path(__file__).parent.parent.as_posix() + '/logs')
# create the directory if not available
pathlib.Path(log_dir_path).mkdir(parents = True, exist_ok = True)
log_file_path = pathlib.Path(log_dir_path + f'/{log_file}')
file_handler = logging.FileHandler(log_file_path)
file_handler.setFormatter(formatter)
infologger.addHandler(file_handler)
if __name__ == "__main__" :
logging.info('Testing log')
日志文件快照
数据预处理
我们使用的数据集几乎是干净的,但在正式实践中,情况并非如此。我们需要处理大量杂乱的数据。所以要做好准备!无论如何,最主要的问题是数据集的不平衡,我们需要使其平衡,否则可能会对我们造成很大的挑战。为了解决这个问题,我使用了超采样技术 SMOTE(合成少数群体超采样技术)来平衡数据集,避免模型中出现任何偏差。它在内部使用 K 近邻算法生成合成数据。
import yaml
import pathlib
import pandas as pd
from src.logger import infologger
from sklearn.model_selection import train_test_split
from imblearn.over_sampling import SMOTE
infologger.info("*** Executing: make_dataset.py ***")
# load data
def load_data(file_path: str) -> pd.DataFrame :
try :
df = pd.read_csv(file_path)
except Exception as e :
infologger.info(f'there\'s some issue while loading data [check load_data()]. exc: {e}')
else :
infologger.info(f'data loaded from {file_path}')
return df
# perform basic preprocessing
def preprocess_data(data: pd.DataFrame) -> pd.DataFrame :
try :
data = data.drop('Id', axis = 1)
except Exception as e :
infologger.info(f'there\'s some issue while preprocessig the data [check preprocess_data()]. exc: {e}')
else :
infologger.info('data preprocesed successfully')
return data
def oversampling(X: pd.DataFrame, y: pd.Series, target: str, seed: int) -> pd.DataFrame :
try :
smote = SMOTE(random_state = seed)
X_res, y_res = smote.fit_resample(X, y)
except Exception as e :
infologger.info(f'there\'s an issue while performing over-sampling [check oversampling()]. exc: {e}')
else :
X_res[target] = y_res
infologger.info('data oversampled using SMOTE!')
return X_res
# split the data
def split_data(test_split: int, seed: int, data: pd.DataFrame) -> pd.DataFrame :
try :
train, test = train_test_split(data, test_size = test_split, random_state = seed)
except Exception as e :
infologger.info(f'there\'s some issue while spliting the data [check split_data()]. exc: {e}')
else :
infologger.info(f'data splited with test_split: {test_split} & seed: {seed}')
return train, test
# save the data
def save_data(path: str, train: pd.DataFrame, test: pd.DataFrame) -> None :
try :
train.to_csv(f'{path}/train.csv', index = False)
test.to_csv(f'{path}/test.csv', index = False)
except Exception as e :
infologger.info(f'there\'s some issue while saving the data [check save_data()]. exc: {e}')
else :
infologger.info(f'training data saved at {path}')
infologger.info(f'testing data saved at {path}')
def main() -> None :
curr_dir = pathlib.Path(__file__)
home_dir = curr_dir.parent.parent.parent
params_file_loc = f'{home_dir.as_posix()}/params.yaml'
try :
params = yaml.safe_load(open(params_file_loc, encoding = 'utf8'))
except Exception as e :
infologger.info(f'there\'s some issue while loading params.yaml [check main()]. exc: {e}')
else :
parameters = params['make_dataset']
TARGET = params['base']['target']
# loading the data from data/raw dir
file_path = f'{home_dir.as_posix()}{params["load_dataset"]["raw_data"]}/{params["load_dataset"]["file_name"]}.csv'
data = preprocess_data(load_data(file_path))
# oversampling() ko data de or ye new aage pass kr
X = data.drop(columns = [TARGET])
y = data[TARGET]
data_res = oversampling(X = X, y = y, target = TARGET, seed = parameters['res_seed'])
train, test = split_data(parameters['test_split'], parameters['seed'], data = data_res)
path = f'{home_dir.as_posix()}{parameters["processed_data"]}'
save_data(path = path, train = train, test = test)
infologger.info('program terminated normally!')
if __name__ == "__main__" :
infologger.info('make_dataset.py as __main__')
main()
特征工程
特征工程在构建高效的机器学习模型中扮演着非常关键的角色。我创建了一些信息丰富的相关特征,它们可以提高机器学习模型的性能。精心设计的特征可以捕捉数据中的重要模式和关系,从而实现更准确的预测。我使用了一些领域知识并生成了一些模型特征。
import pathlib
import yaml
import numpy as np
import pandas as pd
from src.logger import infologger
infologger.info("*** Executing: build_features.py ***")
# writing import after infologger to log the info precisely
from src.data.make_dataset import load_data
# build augmented features
def feat_eng(df: pd.DataFrame, name: str = 'default') -> pd.DataFrame :
try :
df.columns = df.columns.str.replace(' ', '_')
df['total_acidity'] = df['fixed_acidity'] + df['volatile_acidity'] + df['citric_acid']
df['acidity_to_pH_ratio'] = df['total_acidity'] / df['pH']
df['free_sulfur_dioxide_to_total_sulfur_dioxide_ratio'] = df['free_sulfur_dioxide'] / df['total_sulfur_dioxide']
df['alcohol_to_acidity_ratio'] = df['alcohol'] / df['total_acidity']
df['residual_sugar_to_citric_acid_ratio'] = df['residual_sugar'] / df['citric_acid']
df['alcohol_to_density_ratio'] = df['alcohol'] / df['density']
df['total_alkalinity'] = df['pH'] + df['alcohol']
df['total_minerals'] = df['chlorides'] + df['sulphates'] + df['residual_sugar']
# Cleaning inf or null values that may result from the operations above
df = df.replace([np.inf, -np.inf], 0)
df = df.dropna()
except Exception as e :
infologger.info(f'there\'s some issue while performing feature eng [check feat_eng()]. exc: {e}')
else :
infologger.info(f'features generated successfully - {name}')
return df
# save the data
def save_data(path: str, train: pd.DataFrame, test: pd.DataFrame) -> None :
try :
train.to_csv(f'{path}/extended_train.csv', index = False)
test.to_csv(f'{path}/extended_test.csv', index = False)
except Exception as e :
infologger.info(f'there\'s some issue while saving the data [check save_data()]. exc: {e}')
else :
infologger.info(f'features generated for training & testing data. saved at loc {path}')
def main() -> None :
curr_dir = pathlib.Path(__file__)
home_dir = curr_dir.parent.parent.parent
params_file_loc = f'{home_dir.as_posix()}/params.yaml'
try :
params = yaml.safe_load(open(params_file_loc, encoding = 'utf8'))
except Exception as e :
infologger.info(f'there\'s some issue while loading params.yaml [check main()]. exc: {e}')
else :
parameters = params['build_features']
processed_data = params['make_dataset']['processed_data']
# load the data
train_data_loc = f'{home_dir.as_posix()}{processed_data}/train.csv'
test_data_loc = f'{home_dir.as_posix()}{processed_data}/test.csv'
train_data, test_data = load_data(train_data_loc), load_data(test_data_loc)
# save the data
path = f'{home_dir.as_posix()}{parameters["extended_data"]}'
save_data(path, feat_eng(train_data, 'training_data'), feat_eng(test_data, 'testing_data'))
infologger.info('program terminated normally!')
if __name__ == "__main__" :
infologger.info('build_features as __main__')
main()
模型训练
完成数据预处理和特征工程后,我们就可以进行模型训练实验,并建立一个性能优越的模型。
下面添加了使用 mlflow 跟踪模型训练实验的小代码片段。添加了注释以便更好地理解。
# address where MLflow tracking server will be hosted
remote_server_uri = http://localhost:5000
# give any name to experiment
exp_name = 'demo_1'
# setting remote server uri
mlflow.set_tracking_uri(remote_server_uri)
# setting experiment name
mlflow.set_experiment(experiment_name = exp_name)
# description for experiment
experiment_description = ('training and keeping eagle eye on random forest classifier models performance. Obj is to predict the quality of wine based on various physicochemical features')
# adding description to the experiment for better understanding (recommanded)
mlflow.set_experiment_tag("mlflow.note.content", experiment_description)
# below description will be for each run
with mlflow.start_run(description = 'Using random forest classifier algo - by ronil') :
# logging the model prarmeters
mlflow.log_params({"n_estimator": ..., "criterion": ..., "max_depth": ..., "seed": ...})
# logging the metrics
mlflow.log_metrics({"accuracy": ..., "precision": ..., "recall": ..., "roc_score": ...})
# logging the current run's model
log_model(model_object, "model")
# logging confusion matrix img as an artifacts
mlflow.log_artifact(filename, 'confusion_matrix')
# setting tags to each run
mlflow.set_tags({'project_name': 'wine-quality', 'project_quarter': 'Q1-2024', 'ml_model' : 'RFC'})
实际模型训练代码片段:
import pathlib
import yaml
import joblib
import mlflow
import typing
import numpy as np
import pandas as pd
from mlflow.sklearn import log_model
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
from sklearn.base import BaseEstimator
from src.logger import infologger
infologger.info('*** Executing: train_model.py ***')
# writing import after infologger to log the info precisely
from src.visualization import visualize
from src.data.make_dataset import load_data
def train_model(training_feat: np.ndarray, y_true: pd.Series, n_estimators: int, criterion: str, max_depth: int, min_samples_split: int, min_samples_leaf: int, random_state: int, yaml_file_obj: typing.IO) -> dict :
try :
model = RandomForestClassifier(n_estimators = n_estimators, criterion = criterion, max_depth = max_depth, min_samples_split = min_samples_split,
min_samples_leaf = min_samples_leaf, random_state = random_state)
model.fit(training_feat, y_true)
except Exception as e :
infologger.info(f'there\'s an issue while training model [check train_model()]. exc: {e}')
else :
infologger.info(f'trained {type(model).__name__} model')
y_pred = model.predict(training_feat)
y_pred_prob = model.predict_proba(training_feat)
accuracy = metrics.balanced_accuracy_score(y_true, y_pred)
precision = metrics.precision_score(y_true, y_pred, zero_division = 1, average = 'macro')
recall = metrics.recall_score(y_true, y_pred, average = 'macro')
roc_score = metrics.roc_auc_score(y_true, y_pred_prob, average = 'macro', multi_class = 'ovr')
return {'model': model, 'y_pred': y_pred, 'params': {"n_estimator": n_estimators, "criterion": criterion,
"max_depth": max_depth, "seed": random_state},
'metrics': {"accuracy": accuracy, "precision": precision, "recall": recall,
"roc_score": roc_score}}
def save_model(model: BaseEstimator, model_dir: str) -> None :
try :
joblib.dump(model, f'{model_dir}/model.joblib')
except Exception as e :
infologger.info(f'there\'s an issue while saving the model [check save_model(). exc: {e}')
else :
infologger.info(f'model saved at {model_dir}')
def main() -> None :
curr_path = pathlib.Path(__file__)
home_dir = curr_path.parent.parent.parent
params_loc = f'{home_dir.as_posix()}/params.yaml'
plots_dir = f'{home_dir.as_posix()}/plots/training'
try :
params = yaml.safe_load(open(params_loc, encoding = 'utf8'))
except Exception as e :
infologger.info(f'there\'s an issue while loading params.yaml [check main()]. exc: {e}')
else :
parameters = params['train_model']
TARGET = params['base']['target']
train_data = f"{home_dir.as_posix()}{params['build_features']['extended_data']}/extended_train.csv"
model_dir = f"{home_dir.as_posix()}{parameters['model_dir']}"
pathlib.Path(model_dir).mkdir(parents = True, exist_ok = True)
data = load_data(train_data)
X_train = data.drop(columns = [TARGET]).values
Y = data[TARGET]
details = train_model(X_train, Y, parameters['n_estimators'], parameters['criterion'], parameters['max_depth'], min_samples_leaf = parameters['min_samples_leaf'],
min_samples_split = parameters['min_samples_split'], random_state = parameters['random_state'], yaml_file_obj = params)
filename = visualize.conf_matrix(Y, details['y_pred'], labels = details['model'].classes_, path = plots_dir, params_obj = params)
mlflow_config = params['mlflow_config']
remote_server_uri = mlflow_config['remote_server_uri']
exp_name = mlflow_config['trainingExpName']
mlflow.set_tracking_uri(remote_server_uri)
mlflow.set_experiment(experiment_name = exp_name)
# adding experiment description
experiment_description = ('training and keeping eagle eye on random forest classifier models performance. Obj is to predict the quality of wine based on various physicochemical features')
mlflow.set_experiment_tag("mlflow.note.content", experiment_description)
# runs description
with mlflow.start_run(description = 'Using random forest classifier algo - by ronil') :
# logging the prarmeters
mlflow.log_params({"n_estimator": details['params']['n_estimator'], "criterion": details['params']['criterion'],
"max_depth": details['params']['max_depth'], "seed": details['params']['random_state']})
# logging metrics
mlflow.log_metrics({"accuracy": details['metrics']['accuracy'], "precision": details['metrics']['precision'],
"recall": details['metrics']['recall'], "roc_score": details['metrics']['roc_score']})
# loagging the current run's model
log_model(details['model'], "model")
# logging confusion matrix img
mlflow.log_artifact(filename, 'confusion_matrix')
# setting tags to each run
mlflow.set_tags({'project_name': 'wine-quality', 'project_quarter': 'Q1-2024', 'ml_model' : 'RFC'})
save_model(details['model'], model_dir)
infologger.info('program terminated normally!')
if __name__ == '__main__' :
infologger.info('train_model.py as __main__')
main()
注意:在执行任何 mlflow 代码之前,请确保你的 mlflow 服务器已启动并运行,因为 mlflow 服务器是实验跟踪、模型注册和工件存储的中心。当我们在内部运行 mlflow 代码时,它会与 MLflow 服务器通信,以记录参数、指标和工件。如果服务器无法运行,你的代码就无法记录这些细节。模型注册完全依赖于 MLflow 服务器。团队成员之间的协作与共享、管理模型和项目工件等功能将无法实现。如果服务器不运行,我们的 MLflow 代码就无法访问这些基本功能。
开启服务器
# command to turn the server up
mlflow server --backend-store-uri sqlite:///mlflow.db
--default-artifact-root ./artifacts
--host localhost
-p 5000
Drill Down 服务器命令 :
- mlflow server:启动服务器的命令
- backend-store-uri sqlite:///mlflow.db:SQLite 作为后端存储,用于存储实验、运行、参数、指标和标签的元数据。现在我们使用 SQLite 作为后端存储,但一旦开始部署,我们就可以使用 Azure SQL Database、Amazon RDS 或任何其他云服务提供商。
- default-artifact-root ./artifacts:将模型、绘图和其他文件等工件存储在 artifacts 目录中。
- host localhost:mlflow 服务器所在主机
- p 5000 : 端口
注意:服务器启动后,请访问 http://localhost:5000
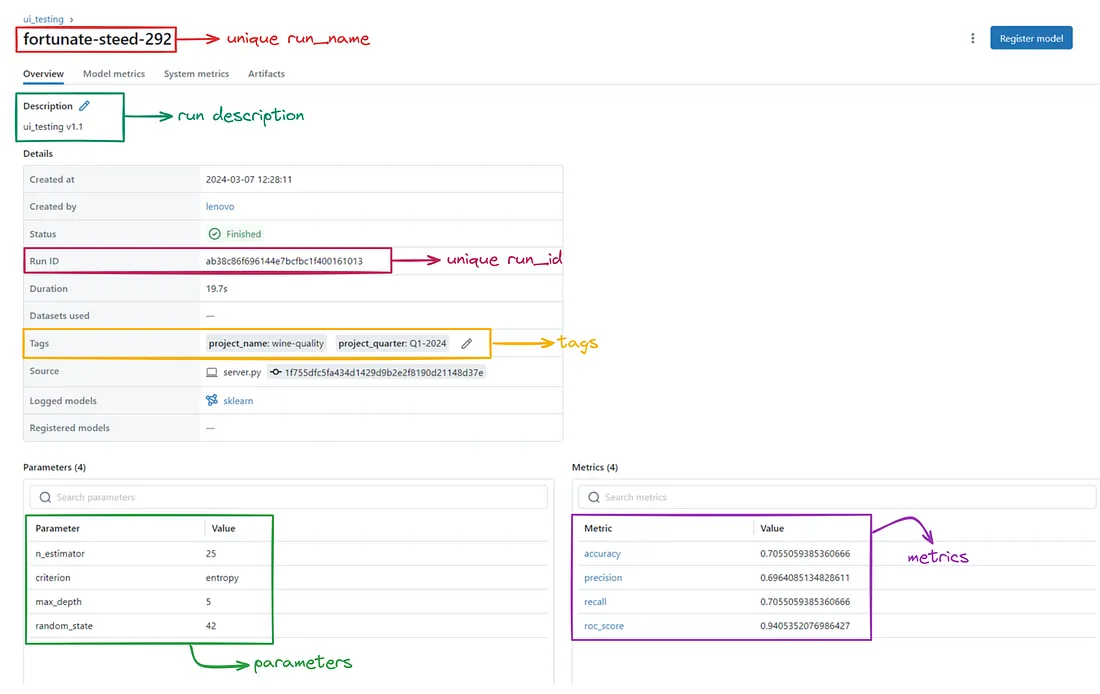
实验与运行
实验是一个容纳运行集合的容器,这些实验与特定的机器学习任务相关。实验有名称、ID、描述和标签。要创建实验,我们可以使用 MLflow 跟踪 API 或 MLflow UI。
运行是实验中模型训练或评估过程的执行。它是使用特定参数、数据或代码训练/调整模型的单次迭代。每个运行都有唯一的名称、运行 ID、描述和标签。运行可捕获元数据,如参数、度量、工件和绘图。要创建运行,我们可以使用 MLflow 跟踪 API 或 MLflow UI。
下面添加了 MLflow UI 的实验和运行部分快照,以便更好地理解。
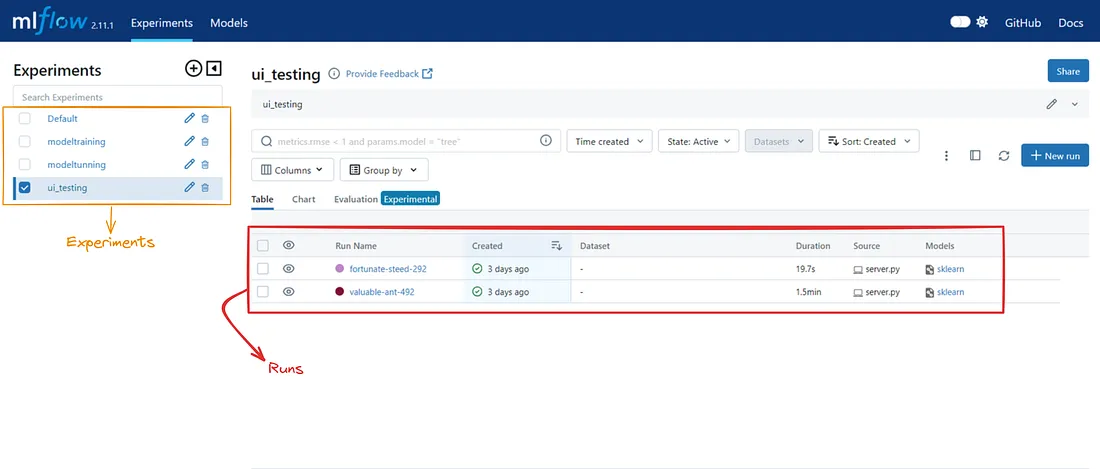
每次运行都会显示以下详细信息。
模型调整
模型训练可能需要尝试各种算法/参数组合,以优化模型,但很少有模型一开箱就能达到最佳性能。这时就需要进行模型调整,它在内部使用统计技术来获取超参数,从而优化模型性能。
我使用 Hyperopt(超参数优化技术)对模型进行了微调。
import mlflow
import yaml
import pathlib
import pandas as pd
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier
from hyperopt import fmin, tpe, hp, STATUS_OK, Trials
from hyperopt.pyll import scope
from src.data.make_dataset import load_data
from functools import partial
from src.logger import infologger
infologger.info('*** Executing: tune_model.py ***')
from src.visualization import visualize
def objective(params: dict, yaml_obj: dict, x_train: pd.DataFrame, y_train: pd.Series, x_test: pd.DataFrame, y_test: pd.Series, plots_dir: str) -> dict :
try :
mlflow_config = yaml_obj['mlflow_config']
remote_server_uri = mlflow_config['remote_server_uri']
exp_name = mlflow_config['tunningExpName']
mlflow.set_tracking_uri(remote_server_uri)
mlflow.set_experiment(experiment_name = exp_name)
# adding experiment description
experiment_description = ('optimizing the hyperparameters of a machine learning model using Hyperopt')
mlflow.set_experiment_tag("mlflow.note.content", experiment_description)
except Exception as e :
infologger.info(f'exception occured while intializing mlflow exp [check objective()]. exc: {e}')
else :
try :
model = RandomForestClassifier(**params)
model.fit(x_train, y_train)
y_pred = model.predict(x_test)
y_pred_prob = model.predict_proba(x_test)
accuracy = metrics.balanced_accuracy_score(y_test, y_pred)
precision = metrics.precision_score(y_test, y_pred, zero_division = 1, average = 'macro')
recall = metrics.recall_score(y_test, y_pred, average = 'macro')
roc_score = metrics.roc_auc_score(y_test, y_pred_prob, average = 'macro', multi_class = 'ovr')
with mlflow.start_run(description = 'tunning RFC also using hyperopt optimization technique') :
mlflow.set_tags({'project_name': 'wine-quality', 'author' : 'ronil', 'project_quarter': 'Q1-2024'})
mlflow.log_params(params)
filename = visualize.conf_matrix(y_test, y_pred, model.classes_, path = plots_dir, params_obj = yaml_obj)
mlflow.log_artifact(filename, 'confusion_matrix')
mlflow.log_metrics({"accuracy": accuracy, "precision": precision, "recall": recall, "roc_score": roc_score})
mlflow.sklearn.log_model(model, 'model')
except Exception as e :
infologger.info(f'got exception while tracking exeriments [check objective()]. exc: {e}')
else :
return {'loss': -accuracy, 'status': STATUS_OK}
def main() -> None :
curr_dir = pathlib.Path(__file__)
home_dir = curr_dir.parent.parent.parent
cm_dir = f'{home_dir.as_posix()}/plots/tunning'
params = yaml.safe_load(open(f'{home_dir.as_posix()}/params.yaml'))
parameters = params['train_model']
TARGET = params['base']['target']
train_data = f"{home_dir.as_posix()}{params['build_features']['extended_data']}/extended_train.csv"
test_data = f"{home_dir.as_posix()}{params['build_features']['extended_data']}/extended_test.csv"
train_data = load_data(train_data)
x_train = train_data.drop(columns = [TARGET]).values
y_train = train_data[TARGET]
test_data = load_data(test_data)
x_test = test_data.drop(columns = [TARGET]).values
y_test = test_data[TARGET]
# hyperopt
additional_params = {'yaml_obj': params, 'x_train': x_train, 'y_train': y_train, 'x_test': x_test, 'y_test': y_test,
'plots_dir': cm_dir}
partial_obj = partial(objective, **additional_params)
# we can take the range as input via params.yaml
search_space = {'n_estimators': hp.randint('n_estimators', 200 - 15) + 15,
'criterion': hp.choice('criterion', ['gini', 'entropy']),
'max_depth': hp.randint('max_depth', 100 - 5) + 5,
'min_samples_split': hp.randint('min_samples_split', 100 - 5) + 5,
'min_samples_leaf': hp.randint('min_samples_leaf', 100 - 10) + 10 }
try :
best_result = fmin(fn = partial_obj,
space = search_space,
algo = tpe.suggest,
max_evals = params['hyperopt']['max_eval'],
trials = Trials())
except Exception as e :
infologger.info(f'exception raised while tunning model using hyperopt [check main()]. exc: {e}')
else :
infologger.info('program terminated normally!')
if __name__ == '__main__' :
main()
infologger.info('tune_model.py as __main__')
# UserWarning: Distutils was imported before Setuptools, but importing Setuptools also replaces the `distutils` module
# in `sys.modules`. This may lead to undesirable behaviors or errors. To avoid these issues, avoid
# using distutils directly, ensure that setuptools is installed in the traditional way (e.g. not an
# editable install), and/or make sure that setuptools is always imported before distutils.
# Solution : So I simply deleted the _distutils_hack and distutils-precedence.pth from the site-packages directory.
# So far so good, though ymmv! My best guess is that those are left behind from some older version of
# setuptools and are not removed when setuptools is updated.
在记录参数和矩阵的同时,我还记录了混淆度量图像作为人工制品。下面我添加了它的实现。
import pathlib
import yaml
import typing
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import joblib
from datetime import datetime
from sklearn.metrics import ConfusionMatrixDisplay, confusion_matrix
from src.logger import infologger
from sklearn.base import BaseEstimator
infologger.info('*** Executing: visualize.py ***')
from src.data.make_dataset import load_data
def load_model(model_dir: str) -> BaseEstimator :
try :
model = joblib.load(model_dir)
except Exception as e :
infologger.info(f'exception raised while loading the model from {model_dir} [check load_model()]. exc: {e}')
else :
infologger.info(f'model loaded successfully from {model_dir}')
return model
def roc_curve() -> None :
pass
def conf_matrix(y_test: pd.Series, y_pred: pd.Series, labels: np.ndarray, path: pathlib.Path, params_obj: typing.IO) -> str :
try :
curr_time = datetime.now().strftime('%d%m%y-%H%M%S')
dir_path = pathlib.Path(f'{path}/confusionMat')
dir_path.mkdir(parents = True, exist_ok = True)
except Exception as e :
infologger.info(f'there\'s an issue in directory [check conf_metrix()]. exc: {e}')
else :
infologger.info('directories are all set!')
try :
cm = confusion_matrix(y_test, y_pred, labels = labels)
disp = ConfusionMatrixDisplay(confusion_matrix = cm, display_labels = labels)
disp.plot(cmap = plt.cm.Blues)
plt.title('Confusion Matrix')
plt.xlabel('Predicted Label')
plt.ylabel('True Label')
filename = f'{dir_path.as_posix()}/{curr_time}.png'
plt.savefig(filename)
plt.close()
except Exception as e :
infologger.info(f'there\'s some issue in ploting confusion metrix [check conf_metrix()]. exc: {e}')
else :
infologger.info(f'confusion metrix saved at [{dir_path}]')
return filename
def main() -> None :
curr_dir = pathlib.Path(__file__)
home_dir = curr_dir.parent.parent.parent
dir_path = f'{home_dir.as_posix()}/plots'
# dir_path.mkdir(parents = True, exist_ok = True)
try :
params = yaml.safe_load(open(f'{home_dir.as_posix()}/params.yaml', encoding = 'utf8'))
except Exception as e :
infologger.info(f'there\'s some issue while loading params.yaml [check main()]. exc: {e}')
else :
data_dir = f"{home_dir.as_posix()}{params['build_features']['extended_data']}/extended_test.csv"
model_dir = f'{home_dir.as_posix()}{params["train_model"]["model_dir"]}/model.joblib'
TARGET = params['base']['target']
test_data = load_data(data_dir)
x_test = test_data.drop(columns = [TARGET]).values
y_test = test_data[TARGET]
model = load_model(model_dir)
labels = model.classes_
try :
y_pred = model.predict(x_test) # return class
except Exception as e :
infologger.info(f'there\'s an issue while prediction [check main()]. exc: {e}')
else :
conf_matrix(y_test, y_pred, labels, dir_path, yaml_file_obj = params)
infologger.info('program terminated normally!')
if __name__ == '__main__' :
main()
缝合工作流程
我们已经介绍了数据收集、数据预处理、特征工程、模型训练和模型调整阶段。这些阶段是机器学习工作流程中非常常见的步骤。我们可以自动执行这些阶段,以实现机器学习实验和数据处理工作流的可重现性和版本化。让我们创建一个 DVC 管道来高效执行这些阶段。
stages:
load_dataset:
cmd: python ./src/data/load_dataset.py
deps:
- ./src/data/load_dataset.py
params:
- load_dataset.drive_link
- load_dataset.raw_data
- load_dataset.file_name
outs:
- .${load_dataset.raw_data}/${load_dataset.file_name}.csv
make_dataset:
cmd: python ./src/data/make_dataset.py
deps:
- ./src/data/make_dataset.py
- .${load_dataset.raw_data}/${load_dataset.file_name}.csv
params:
- load_dataset.raw_data
- load_dataset.file_name
- make_dataset.test_split
- make_dataset.seed
- make_dataset.processed_data
- make_dataset.res_seed
outs:
- .${make_dataset.processed_data}/train.csv
- .${make_dataset.processed_data}/test.csv
build_features:
cmd: python ./src/features/build_features.py
deps:
- ./src/features/build_features.py
- .${make_dataset.processed_data}/train.csv
- .${make_dataset.processed_data}/test.csv
params:
- make_dataset.processed_data
- build_features.extended_data
outs:
- .${build_features.extended_data}/extended_train.csv
- .${build_features.extended_data}/extended_test.csv
train_model:
cmd: python ./src/models/train_model.py
deps:
- ./src/models/train_model.py
- .${build_features.extended_data}/extended_train.csv
- .${build_features.extended_data}/extended_test.csv
params:
- build_features.extended_data
- base.target
- train_model.n_estimators
- train_model.criterion
- train_model.random_state
- train_model.max_depth
- train_model.min_samples_leaf
- train_model.min_samples_split
- train_model.model_dir
- mlflow_config.trainingExpName
- mlflow_config.remote_server_uri
outs:
- .${train_model.model_dir}/model.joblib
tune_model:
cmd: python ./src/models/tune_model.py
deps:
- ./src/models/tune_model.py
- .${build_features.extended_data}/extended_train.csv
- .${build_features.extended_data}/extended_test.csv
params:
- build_features.extended_data
- base.target
- mlflow_config.tunningExpName
- mlflow_config.remote_server_uri
- hyperopt.max_eval
将此 dvc.yaml 文件放入根目录,然后在 bash/shell 中运行 dvc repro 命令。
所有配置参数都存储在 params.yaml 中,这样就可以更方便地管理和更新配置,而无需直接修改代码。下面是 params.yaml 的一个片段。
base:
project_name: mlflow-testing
target: quality
load_dataset:
drive_link: https://drive.google.com/file/d/1FCVWcNyBX6tFYjfORhnAvIwS8I8zV92j/view?usp=sharing
raw_data: /data/raw
file_name: wineq
make_dataset:
test_split: 0.25
seed: 41
processed_data: /data/processed
res_seed: 42
build_features:
extended_data: /data/extended
mlflow_config:
# artifacts_dir: artifacts
trainingExpName: modeltraining
tunningExpName: modeltunning
remote_server_uri: http://localhost:5000 # 127.0.0.1
mlflow_tracking_uri: sqlite:///mlflow.db
cmd: mlflow server --backend-store-uri sqlite:///mlflow.db --default-artifact-root ./artifacts --host localhost -p 5000
reg_model_name: prod_testing
stage: production
train_model:
n_estimators: 50
criterion: gini
max_depth: 50
min_samples_leaf: 30
random_state: 42
min_samples_split: 60
model_dir: /models
hyperopt:
max_eval: 5
使用 Streamlit 调整 ML 模型
这是一种有用而有效的方法,可用于试验模型参数,并快速观察不同参数对模型行为的影响。用户无需在每次更改参数时运行整个 ML 管道,而是可以使用 Streamlit 网络应用程序快速试验不同的参数。此外,模型、参数和指标都会被 MLflow 记录下来,以便日后使用。下面是添加的代码,请仔细阅读。
import mlflow
import pathlib
import yaml
import streamlit as st
from mlflow.tracking import MlflowClient
from mlflow.sklearn import load_model
curr_dir = pathlib.Path(__file__)
home_dir = curr_dir.parent.parent
params = yaml.safe_load(open(f'{home_dir.as_posix()}/params.yaml'))
st.set_page_config(page_title = 'WineQ Prediction',
page_icon = '🦅',
layout = 'centered',
initial_sidebar_state = 'expanded')
# we set the tracking URI globally using mlflow.set_tracking_uri(). When we create the MlflowClient object
# without specifying the tracking_uri parameter explicitly, it automatically uses the tracking URI that we've
# set globally. This approach is more concise.
mlflow.set_tracking_uri(params['mlflow_config']['mlflow_tracking_uri'])
client = MlflowClient()
# Sidebar Info
st.sidebar.title("About Me 🤖")
try :
model = client.get_model_version_by_alias(name = params['mlflow_config']['reg_model_name'], alias = params['mlflow_config']['stage'])
st.sidebar.write(f"#### Model Name\n ```{model.name}```")
st.sidebar.write(f"#### Model Version\n ```version v{model.version}```")
st.sidebar.write(f"#### Current Stage")
for i in model.aliases:
st.sidebar.write(f'```{i}```')
st.sidebar.write(f"#### Run ID\n ```{model.run_id}```")
if 'final_model' not in st.session_state :
with st.spinner('Loading Models') :
st.session_state['final_model'] = load_model(f"models:/{model.name}/{model.version}")
st.sidebar.info('##### Server is Up 🔥')
except :
st.sidebar.warning('##### ⚠️ Model not found')
# Body
st.title("Wine Quality 🍷")
fixed_acidity = st.number_input('Fixed Acidity *',min_value = 4.1, max_value = 16.4, value = None, placeholder = '4.1 <= Fixed Acidity <= 16.4', step = 0.1)
volatile_acidity = st.number_input('Volatile Acidity *', placeholder = '0.5 <= Volatile Acidity <= 1.98', min_value = 0.5, max_value = 1.98, value = None, step = 0.1)
citric_acid = st.number_input('Critic Acid *', placeholder = '0.0 <= Citrix Acid <= 1.5', min_value = 0.0, max_value = 1.5, value = None, step = 0.1)
residual_sugar = st.number_input('Residual Sugar *', placeholder = '0.5 <= Residual Sugar <= 16.0', min_value = 0.5, max_value = 16.0, value = None, step = 0.1)
chlorides = st.number_input('Chloride *', placeholder = '0.008 <= Chloride <= 0.7', min_value = 0.008, max_value = 0.7, value = None, step = 0.001, format = "%.3f")
free_sulfur_dioxide = st.number_input('Free Sulfur Dioxide *', placeholder = '0.7 <= Free Sulfur Dioxide <= 70.0', min_value = 0.7, max_value = 70.0, value = None, step = 0.1)
total_sulfur_dioxide = st.number_input('Total Sulfur Dioxide *', placeholder = '5 <= Total Sulfur Dioxide <= 290', min_value = 5, max_value = 290, value = None, step = 4)
density = st.number_input('Density *', placeholder = '0.85 <= Density <= 1.5', min_value = 0.85, max_value = 1.5, value = None, step = 0.1)
pH = st.number_input('PH *', placeholder = '2.6 <= PH <= 4.5', min_value = 2.6, max_value = 4.5, value = None, step = 0.1)
sulphates = st.number_input('Sulphate *', placeholder = '0.2 <= Sulphate <= 2.5', min_value = 0.2, max_value = 2.5, value = None, step = 0.1)
alcohol = st.number_input('Alcohol *', placeholder = '8 <= Alcohol <= 15', min_value = 8, max_value = 15, value = None, step = 1)
user_input = {'fixed_acidity': fixed_acidity, 'volatile_acidity' : volatile_acidity, 'citric_acid': citric_acid, 'residual_sugar' : residual_sugar,
'chlorides': chlorides, 'free_sulfur_dioxide': free_sulfur_dioxide, 'total_sulfur_dioxide': total_sulfur_dioxide, 'density': density,
'pH': pH, 'sulphates': sulphates, 'alcohol': alcohol}
if fixed_acidity and volatile_acidity and citric_acid and residual_sugar and chlorides and free_sulfur_dioxide and total_sulfur_dioxide and density and\
pH and sulphates and alcohol :
# feature engineering
user_input['total_acidity'] = user_input['fixed_acidity'] + user_input['volatile_acidity'] + user_input['citric_acid']
user_input['acidity_to_pH_ratio'] = (lambda total_acidity, pH : 0 if pH == 0 else total_acidity / pH)(user_input['total_acidity'], user_input['pH'])
user_input['free_sulfur_dioxide_to_total_sulfur_dioxide_ratio'] = (lambda free_sulfur_dioxide, total_sulfur_dioxide : 0 if total_sulfur_dioxide == 0 \
else free_sulfur_dioxide / total_sulfur_dioxide)\
(user_input['free_sulfur_dioxide'], user_input['total_sulfur_dioxide'])
user_input['alcohol_to_acidity_ratio'] = (lambda alcohol, total_acidity : 0 if total_acidity == 0 else alcohol / total_acidity)\
(user_input['alcohol'], user_input['total_acidity'])
user_input['residual_sugar_to_citric_acid_ratio'] = (lambda residual_sugar, citric_acid : 0 if citric_acid == 0 else residual_sugar / citric_acid)\
(user_input['residual_sugar'], user_input['citric_acid'])
user_input['alcohol_to_density_ratio'] = (lambda alcohol, density : 0 if density == 0 else alcohol / density)(user_input['alcohol'], user_input['density'])
user_input['total_alkalinity'] = user_input['pH'] + user_input['alcohol']
user_input['total_minerals'] = user_input['chlorides'] + user_input['sulphates'] + user_input['residual_sugar']
input_data = [[user_input['fixed_acidity'], user_input['volatile_acidity'], user_input['citric_acid'], user_input['residual_sugar'],
user_input['chlorides'], user_input['free_sulfur_dioxide'], user_input['total_sulfur_dioxide'], user_input['density'],
user_input['pH'], user_input['sulphates'], user_input['alcohol'], user_input['total_acidity'], user_input['acidity_to_pH_ratio'],
user_input['free_sulfur_dioxide_to_total_sulfur_dioxide_ratio'], user_input['alcohol_to_acidity_ratio'],
user_input['residual_sugar_to_citric_acid_ratio'], user_input['alcohol_to_density_ratio'], user_input['total_alkalinity'],
user_input['total_minerals']]]
if st.button('Predict ⚙️') :
with st.spinner('Training the model...') :
op = st.session_state['final_model'].predict(input_data)[0]
pred_prob = st.session_state['final_model'].predict_proba(input_data)[0]
st.success(f'Predicted Quality is {op} with {(max(pred_prob) * 100):.2f}% model\'s confidence.')
下面添加了 Streamlit 应用程序的快照。
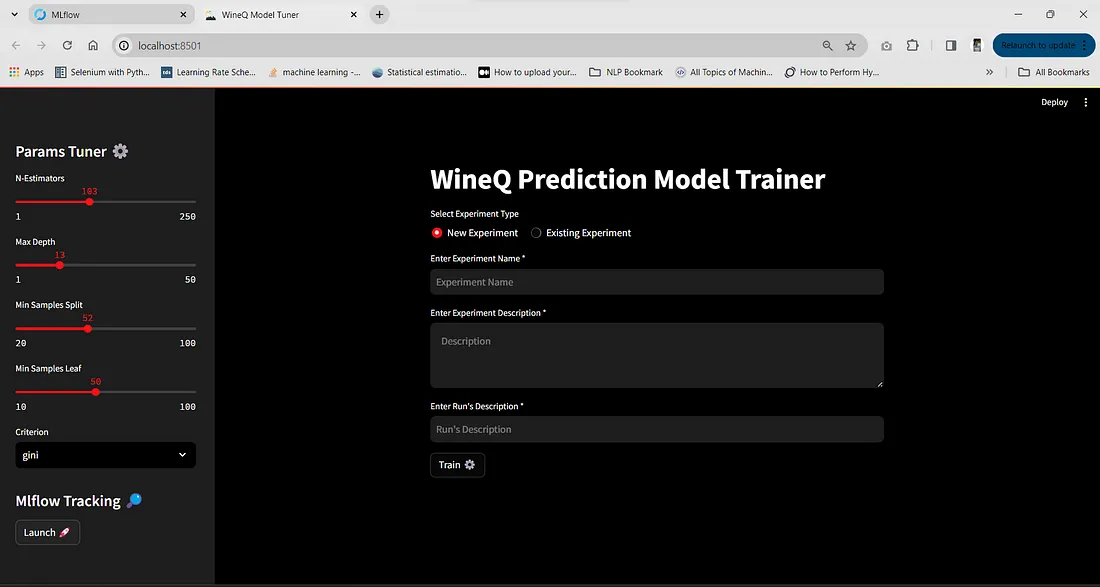
在这里,我添加了创建新实验或使用现有实验来记录模型/运行的选项。此外,我们还可以加入运行和实验描述,这将有助于我们了解每个实验或运行的背景、目标和结果。